|
Our group is probing the structure, dynamics and organization of membranes and membrane proteins from the atomistic level to the systems level. The importance of membrane related processes is reflected in the fact that about 1/3 of the human genome comprises of membrane proteins and almost 60%; of current prescribed drugs target membrane proteins. Unraveling this systems-level cross-talk within the membrane, therefore represents a novel frontier in biology that will open up new strategies to tackling human health and disease.
We have developed frameworks to understand receptor organization at multiple levels. In order to extract general features of receptor organization, we analyse single transmembrane receptors, such as ErbB2, as well as more complex GPCRs. We have also developed models to study membrane topology changes by proteins such as caveolin and LC3. Another aspect of our work is to understand the mechanism of action of membrane active peptides, such as antimicrobial peptides.
1. Membrane Receptor Organization: Association of EGFR in membranes
We are currently studying the association of receptors such as ErbB1 and ErbB2 to understand the organization of these receptors and the driving forces of association. These receptors are central to many signalling pathways and involved in most carcinomas and development defects. The traditional ligand-induced dimerisation paradigm has now been modified to the pre-formed dimer paradigm and further extended to the nano-clusters or signalosome concept ; though none of the models are able to fully explain the specificity and efficiency of these networks.
Refereces:
- D Sengupta@, S J Marrink, 2010, Lipid Mediated Interaction tune the interaction of Glycophorin A helices in Membranes, Phys. Chem. Chem. Phys. 12, 12987-12996
- X Prasanna, P J Praveen, D Sengupta@, 2013,Sequence dependent lipid-mediated effects modulate the dimerization of ErbB2 and its associative mutants, Phys. Chem. Chem. Phys., 15, 19031-19041
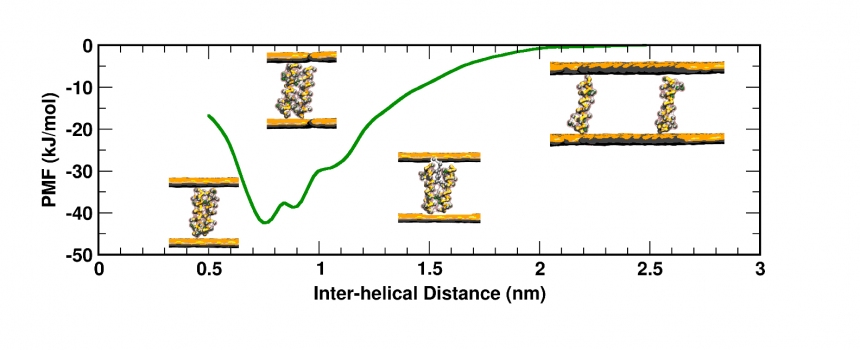
2. GPCR-cholesterol interactions: Serotonin Receptor and the β-2 Adrenergic Receptor
G-protein coupled receptors (GPCRs) represent not just one of the largest families of human proteins but also 50% of the current clinically important drug targets and yet we know very little about the molecular details of their structure, dynamics and organization We are focussing on two receptors, namely the serotonin 1A receptor and the &beta2-adrenergic receptor. Using multi-scale models, we analyse how the membrane environement modulates protein dynamics and function. We delineate whether direct effects such as lipid- and cholesterol-binding or indirect effects such as membrane thickness and topology changes are important in tuning the structural organization of GPCRs.
References
- D Sengupta@ and A Chattopadhyay, 2012, Identification of Cholesterol Binding Sites in the Serotonin1A Receptor, J. Phys. Chem. B, 116, 12991-12996
- Prasanna, A. Chattopadhyay, D Sengupta@, Cholesterol Modulates the Dimer Interface of the &beta2-Adrenergic Receptor via Cholesterol Occupancy Sites, Biophys. J., In Press
3. Lipidated Proteins: Stabilizing or Generating Membrane Curvature?
Lipid modication of cytoplasmic proteins trigger membrane recruitment that is vital for organelle and vesicular dynamics. The main focus of our work is caveolin-1, the most common member of caveolin family and plays multiple roles in cellular signalling, lipid exchange, endocytosis, and intracellular delivery of bacterial toxins and viruses. We aim to understand of the mode of interaction of caveolin-1 peptide with model bilayers. An increased preference for cav-1 peptides for cholesterol-rich membranes is seen in our simulations We also show that the binding of cav-1 peptides to DOPC membranes is the least favourable amongst the model membranes studied.
References
- D Sengupta@, 2012, Cholesterol Modulates the Structure, Binding Modes and Energetics of Caveolin-Membrane Interactions, J. Phys. Chem. B, 116, 14556-14564
4. Antimicrobial Peptide Action
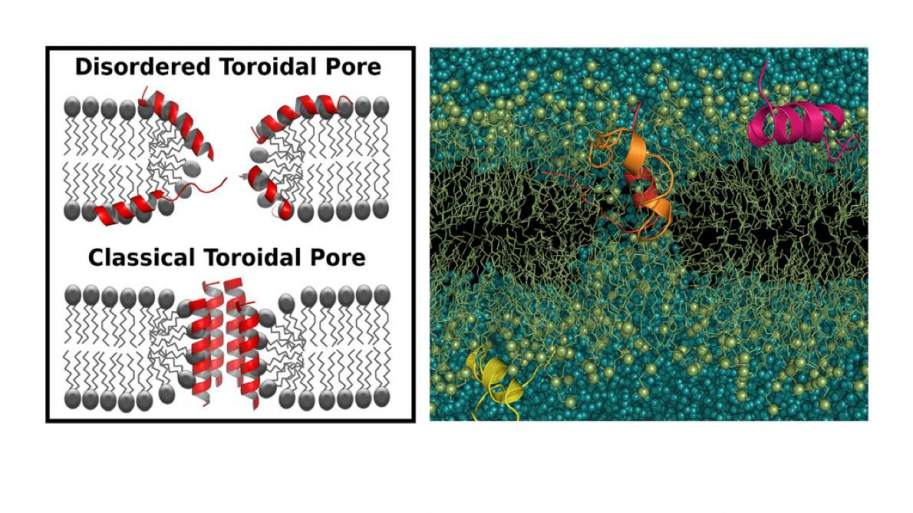
An important focus of our research is the mechanism of action of antimicrobial peptides. We have previously showed the existence of disordered toroidal pores for several of these peptides (See Publications). We have also worked towards developing a framework to effectively combine computer simulations with experimental studies to understand the molecular basis of activity in these peptides. We have studied a cyclic antimicrobial peptide, BPC194, by spectroscopic and computational techniques to infer functionally-important structural features. We have also determined poration propensity and pore size by simulations and successfully related it to measurements from electrophyisiology and fluorescence cross correlation spectroscopy (FCCS) experiments.
We are currently collaborating with Dr. S. Kendurkar, NCL, Pune to develop linear antimicrobial peptides for increased potency against plant pathogens.
Refereces:
- AD Cirac, G Moiset, JT Mika, A Kocer, P Salvador, SJ Marrink, B Poolman and D Sengupta@, 2011, The molecular basis for antimicrobial activity of pore-forming cyclic peptides, Biophys. J., 100, 2422-2431
|



